Common disease genomics in an era of whole genome sequencing at scale
The team study the genetics and genomics of common metabolic disease, focusing on both types of diabetes, obesity and related conditions including obesity associated cancers. To do so, we use a combination of genetic analysis, large international and local datasets and clinical studies involving patients and clinicians. To achieve our goals, we adopt an “industry standard” way of working as a team, using an “Agile” approach to project planning. Local datasets we work with will include a new Hospital biobank, “BioHUG” that will incorporate DNA samples and electronic medical records from >20,000 patients in Geneva.
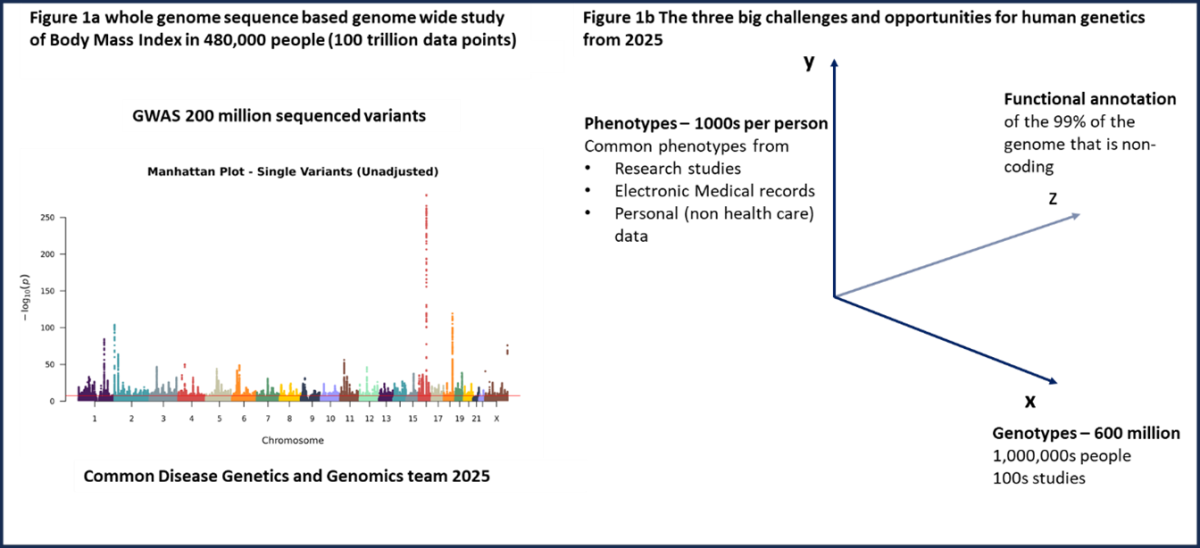
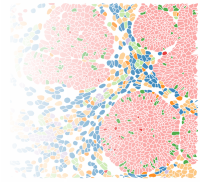
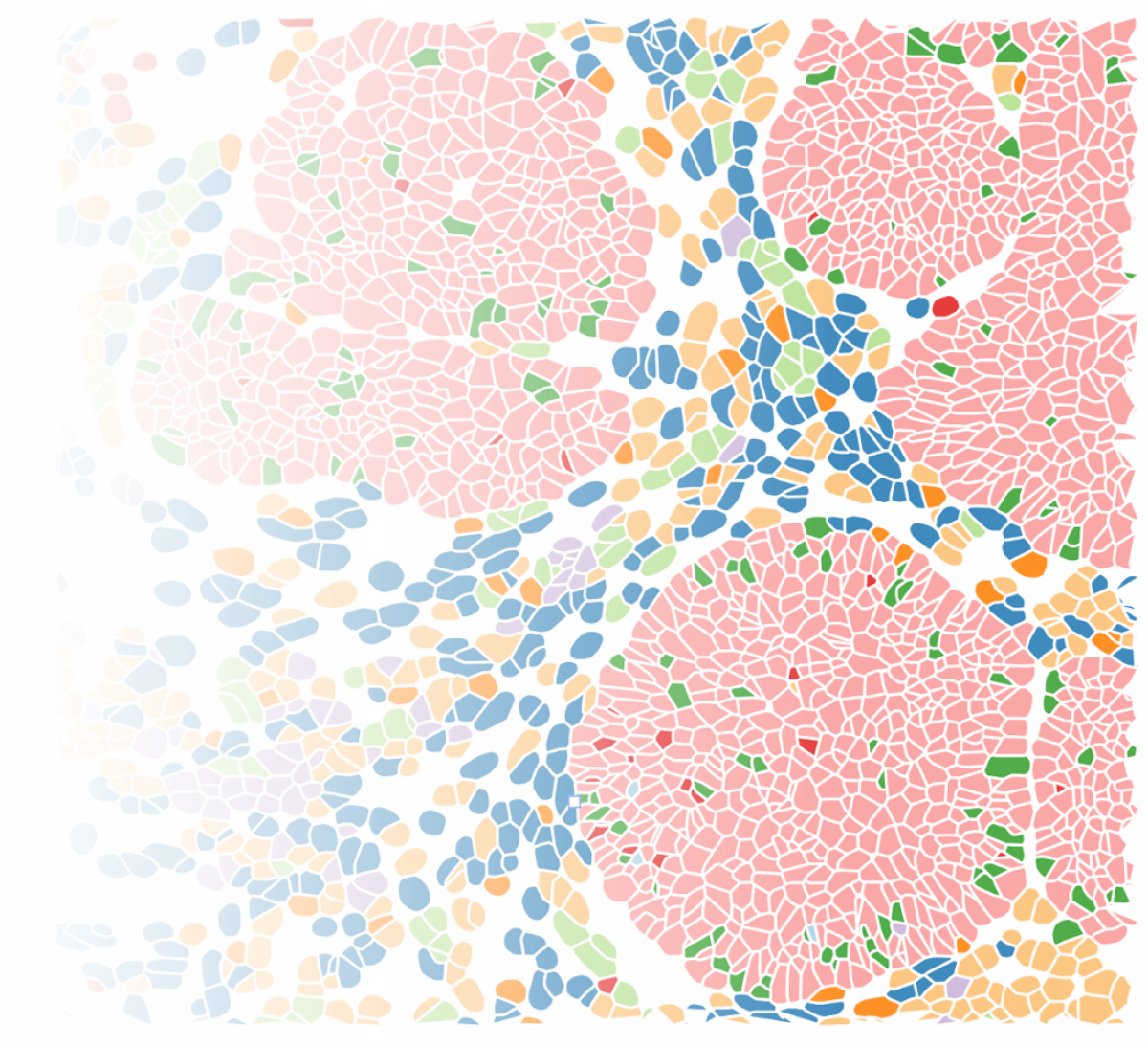
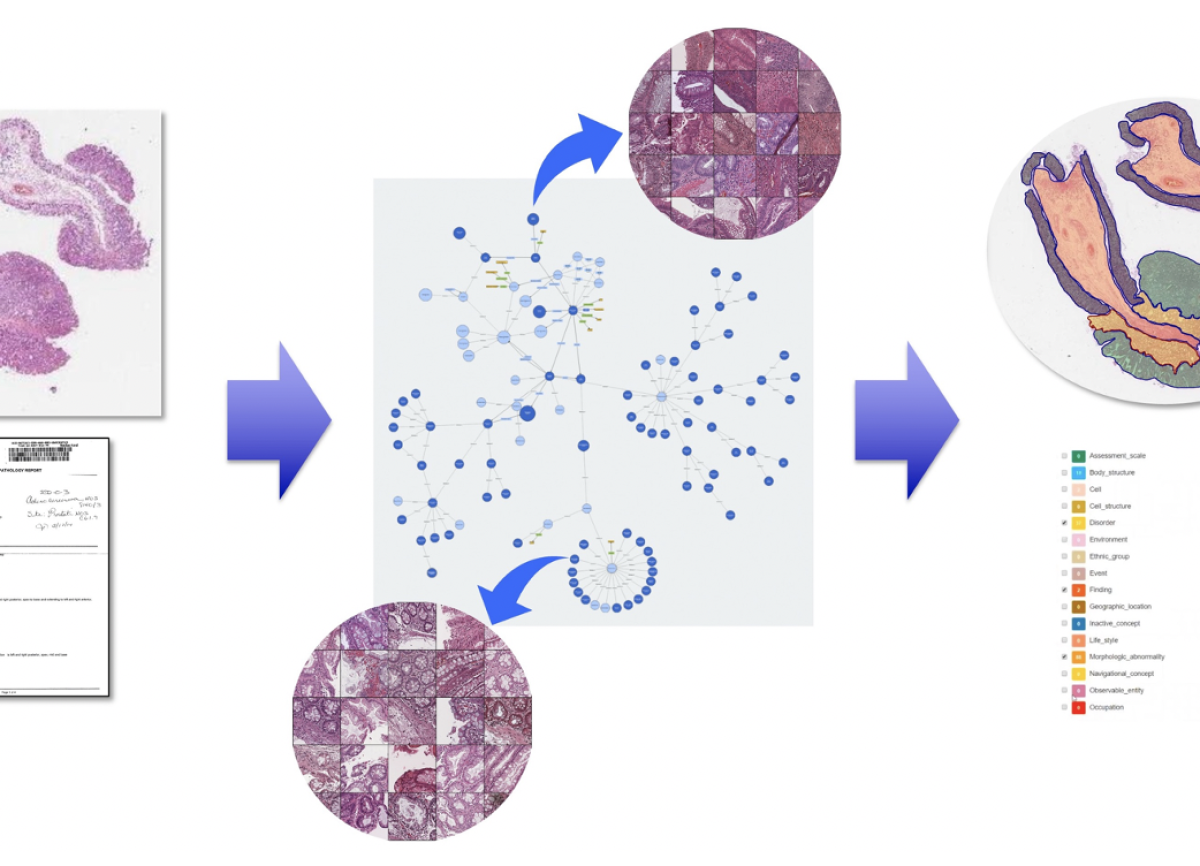
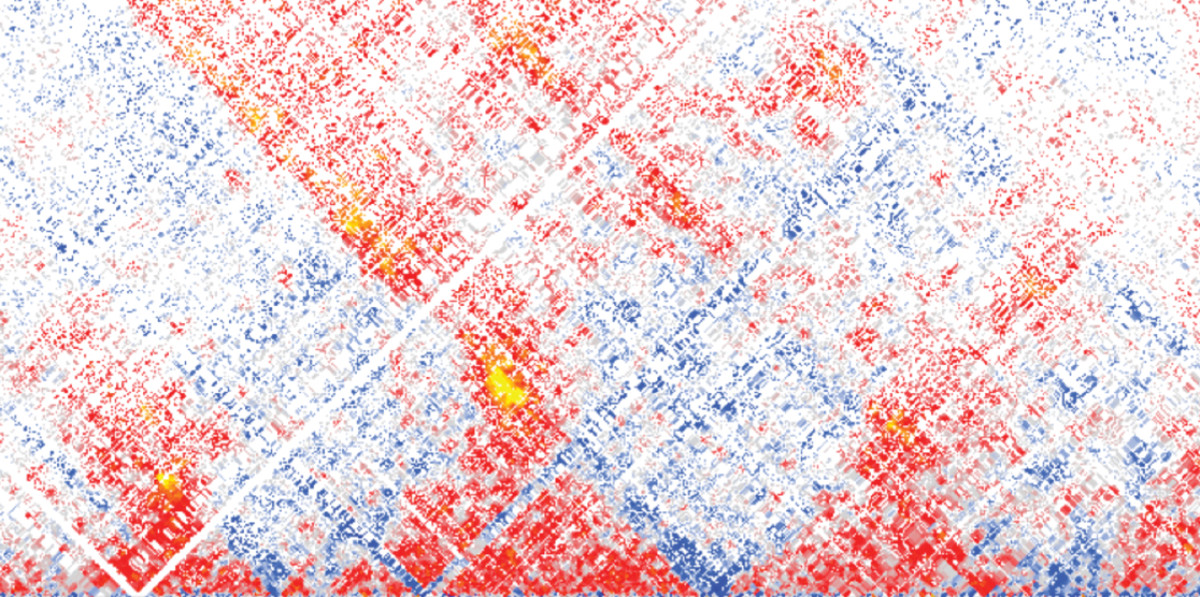
There has never been a more exciting time to study common disease using human genetics. This excitement stems from the advances in technology that means we now work with whole genome sequence (WGS) data in 100’000s of people (based on short read sequencing) or 10,000s of people (based on long read sequencing). This use of sequence level data in the context of common disease is still in its infancy but, for the first time, provides us with the ability to identify rarer variants with relatively large effects on common phenotypes across the whole human genome. The discovery of such alleles will facilitate greater understanding of biology, disease and drug development, through more direct functional and clinical studies, and more opportunities for personalised medicine. The advances in genome sequencing are occurring in parallel to two other broad major advances. First, those in functional genomics, where technologies such as single cell RNA sequencing and chromatin accessibility assays means we will have a much greater understanding of the role of the non-coding genome. Second, in the linkage of human genetic and genomic information to routinely collected medical and health information. These 3 big challenges – genotypes, phenotypes and functional information are illustrated as a notional 3 axis graph in Fig 1b.
Visit our website
Apply now