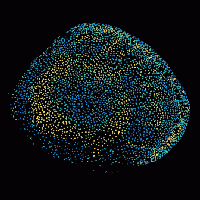
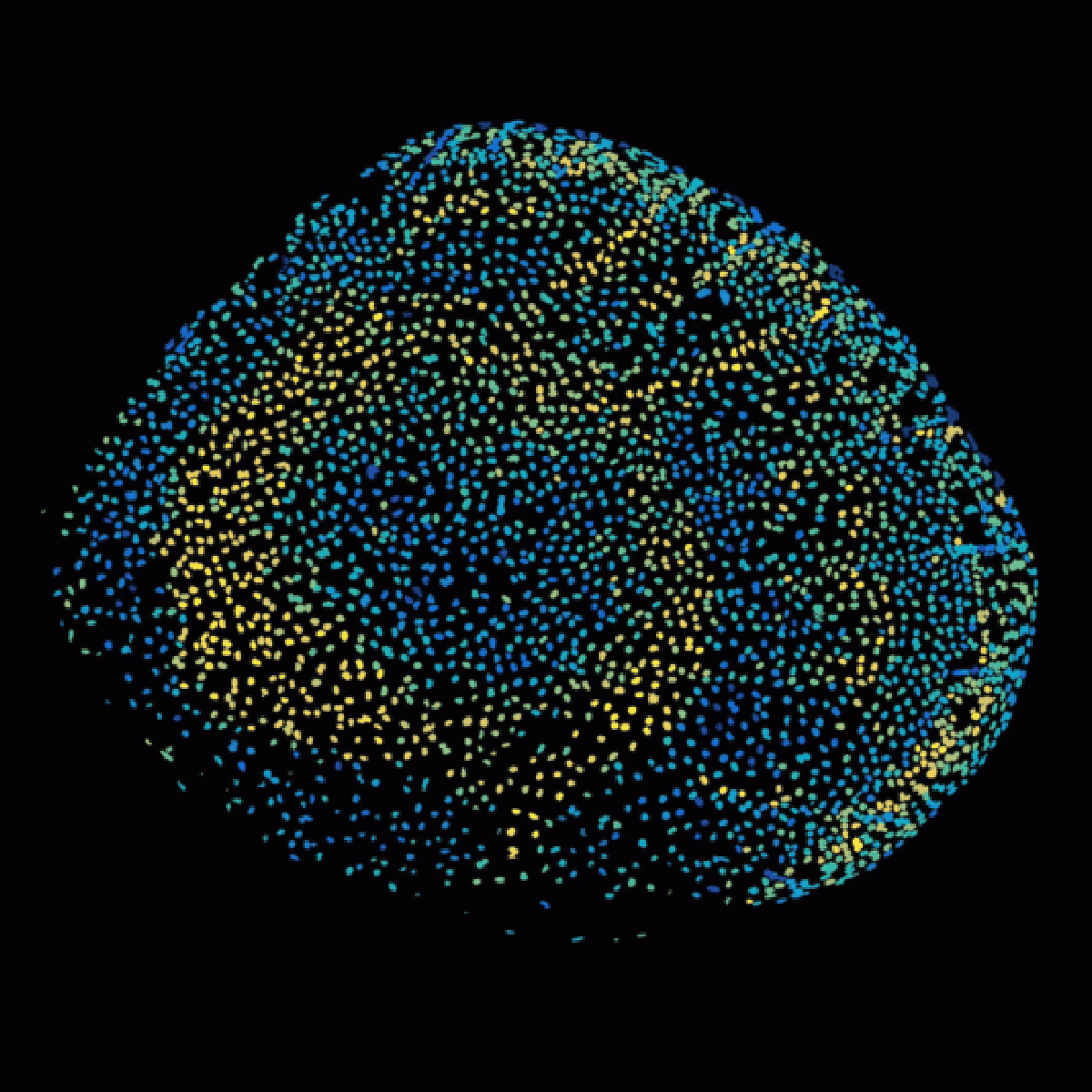
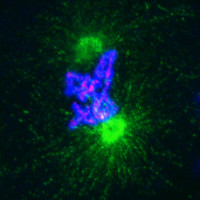
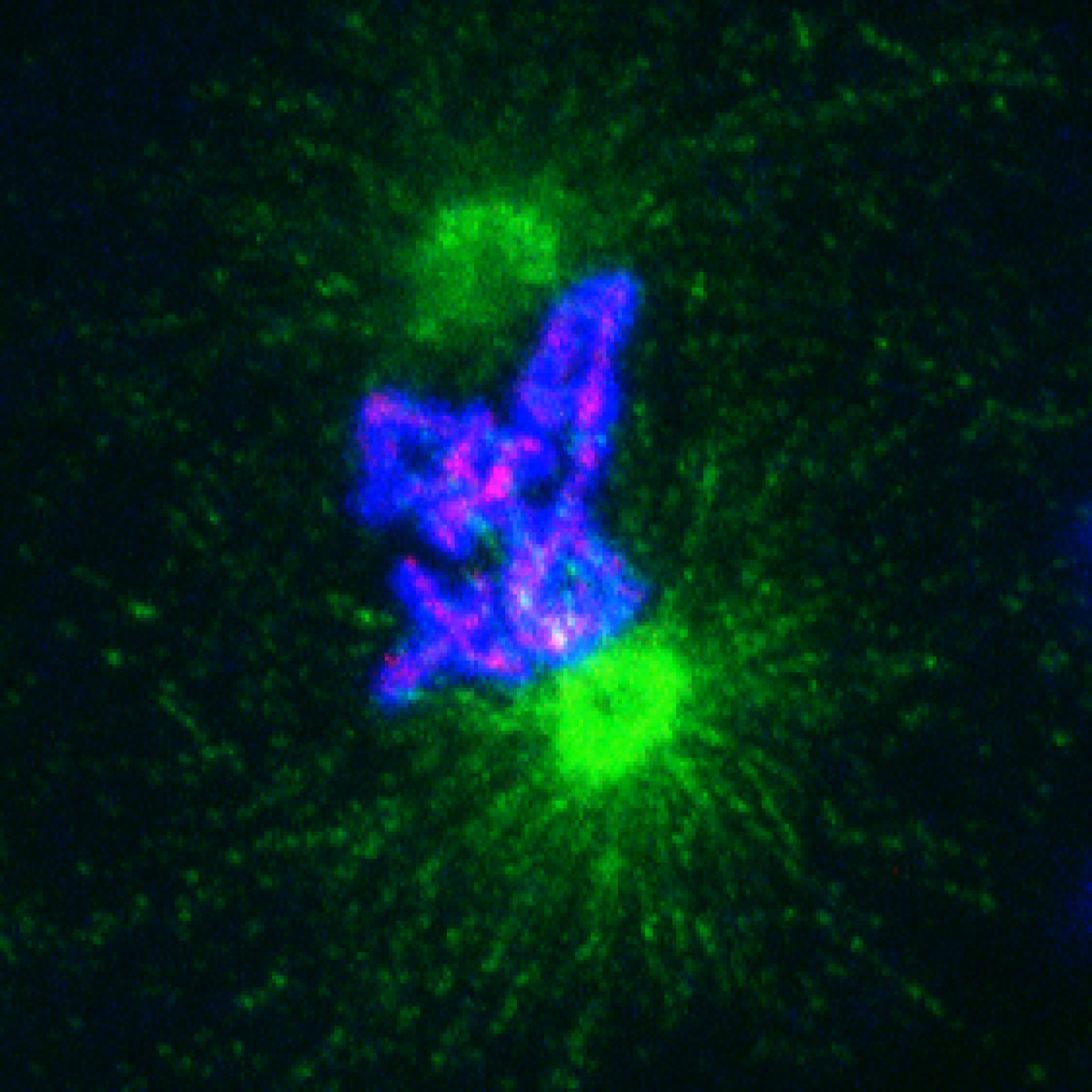
Patterning of the vertebrate skin through mechanical and Turing instabilities
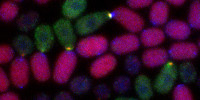
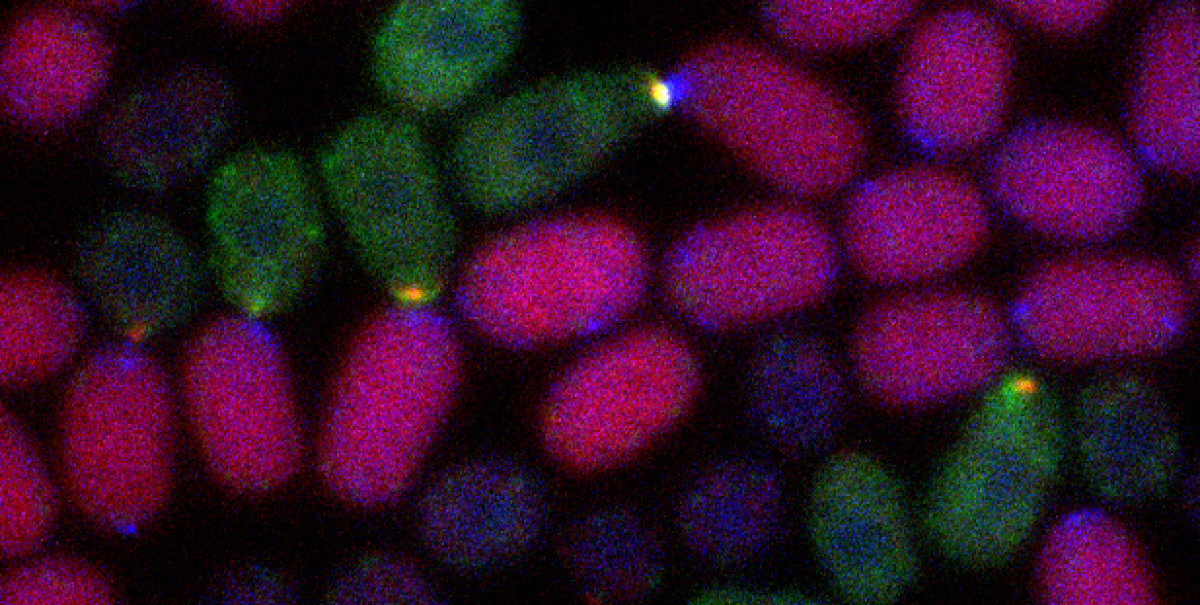
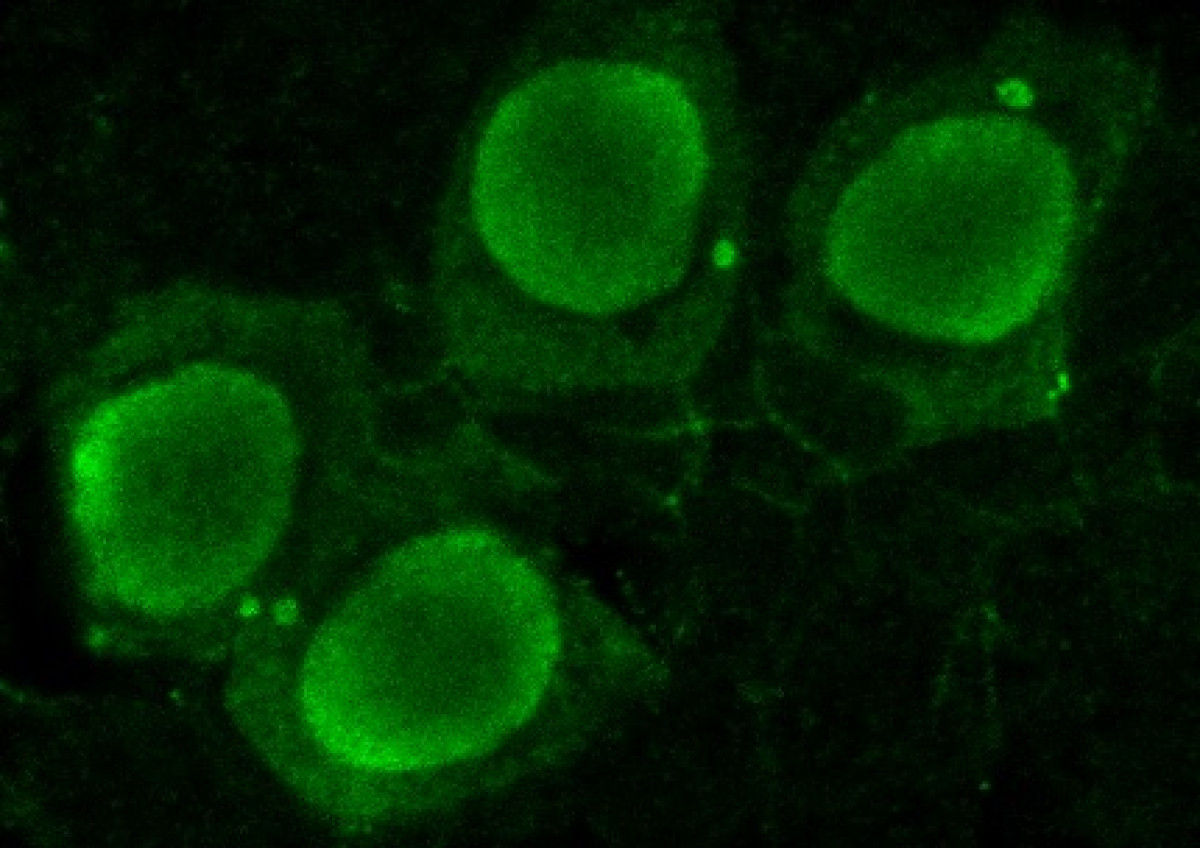
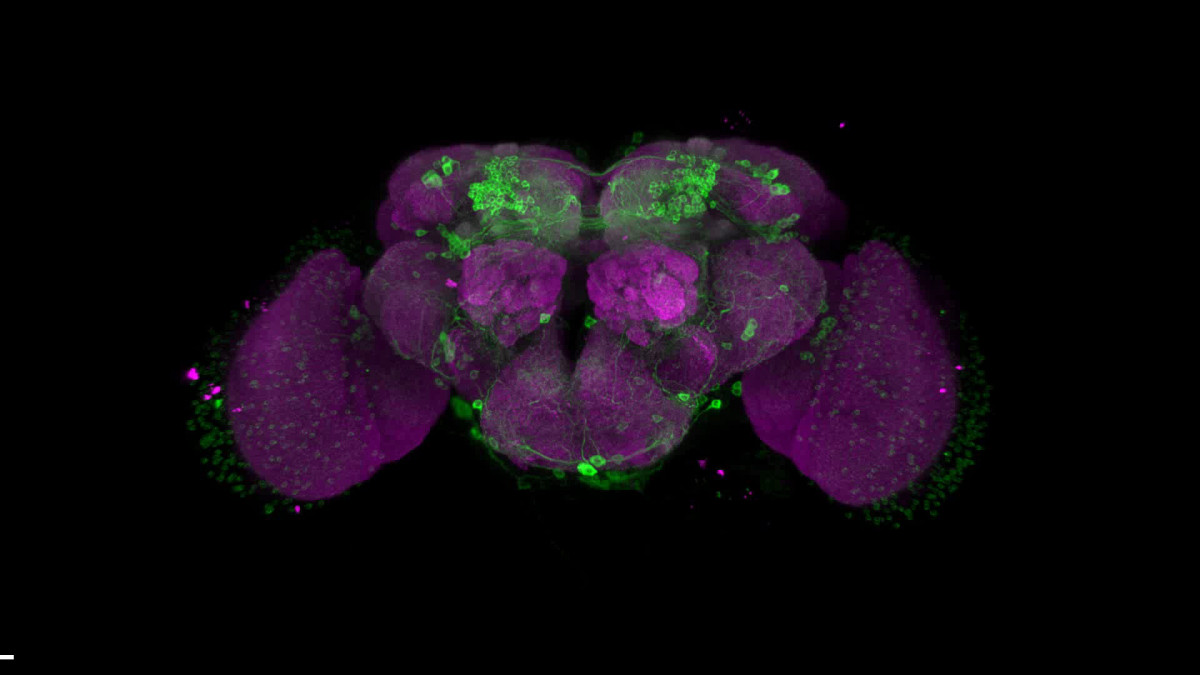
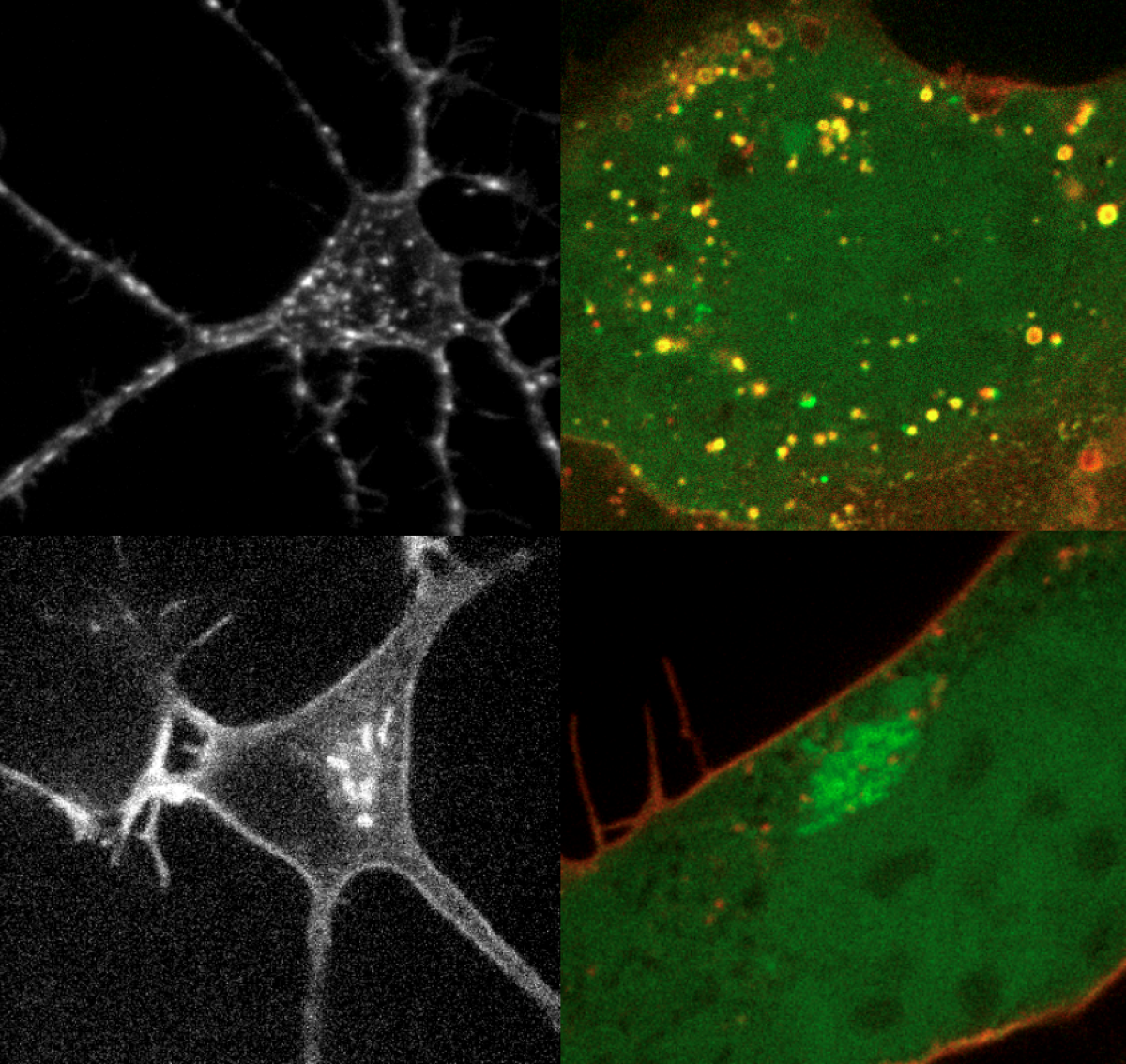
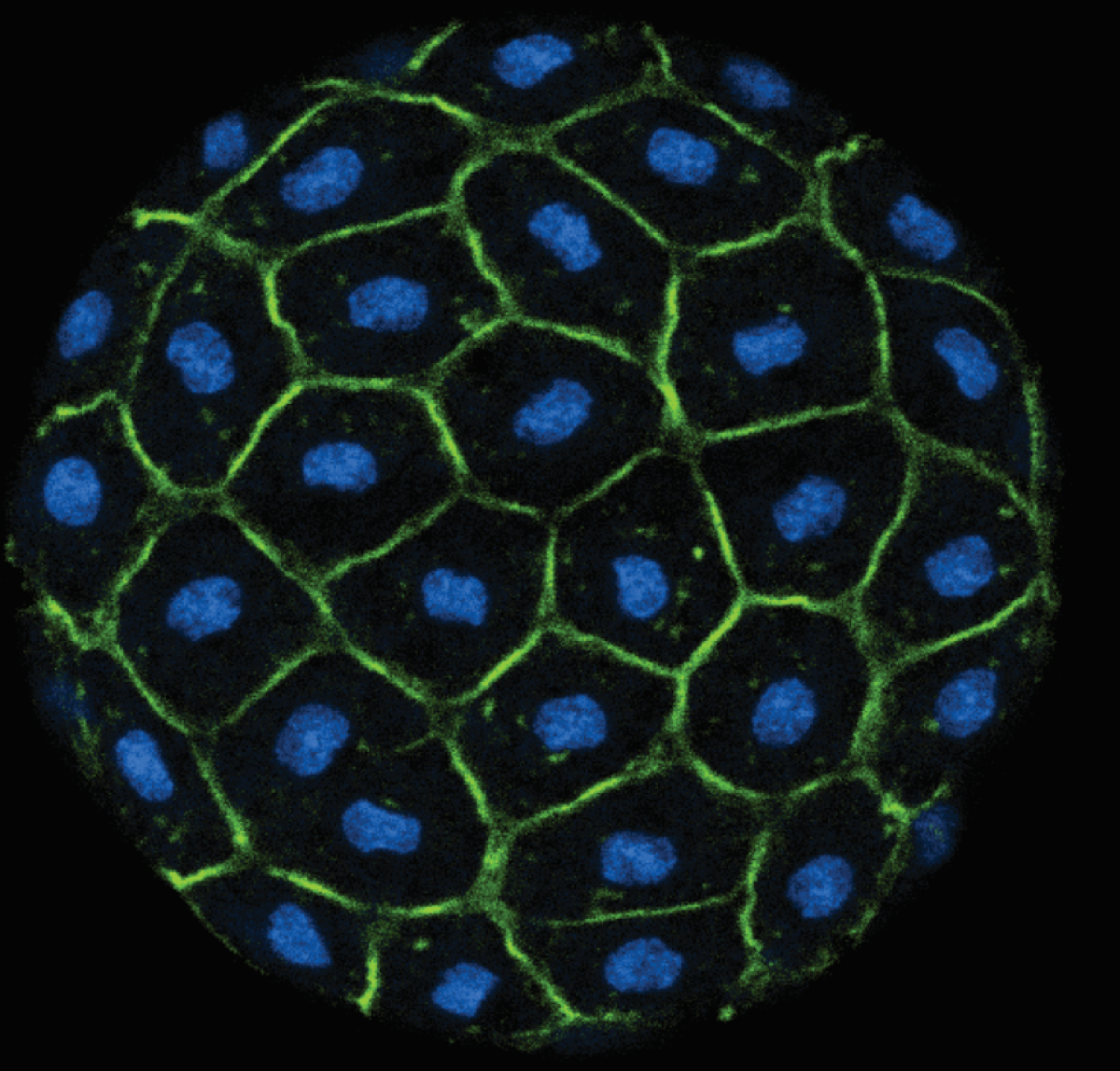
Because discipline-oriented approaches can sometimes restrain imagination, our laboratory includes biologists, physicists and computer scientists, making our research highly multidisciplinary and integrative. One of our main models is the development and evolution of the vertebrate skin, with special emphasis on skin appendages (scales, hairs, spines), skin colours (pigmentary and structural), and skin colour patterns. Our strategy is to tackle these topics from different angles by using multiple techniques (genomics, physical experiments, mathematical modelling and numerical simulations). We work at multiple spatial scales (genomes, cells, tissues, organisms) and use new model species of tetrapods (frogs, snakes, lizards, crocodiles, hedgehogs, tenrecs, spiny mice).
Visit our website
Apply now